Molecular scales on biological membranes: Mass-Sensitive Particle Tracking
New method to determine location and size changes of unlabeled proteins on membranes via their mass
A large proportion of biologically relevant processes take place at membranes. Studying the dynamics of these processes in real time and without disturbing the biological system is still a major methodological challenge. A team led by Petra Schwille, director at the MPI of Biochemistry (MPIB), and Nikolas Hundt from the Ludwig-Maximilians-Universität München (LMU) has now developed a new method for this purpose: Mass-Sensitive Particle Tracking (MSPT). Using MSPT, the movements and reactions of individual unlabeled proteins on biological membranes can be determined solely by their mass. The method has been published in Nature Methods.
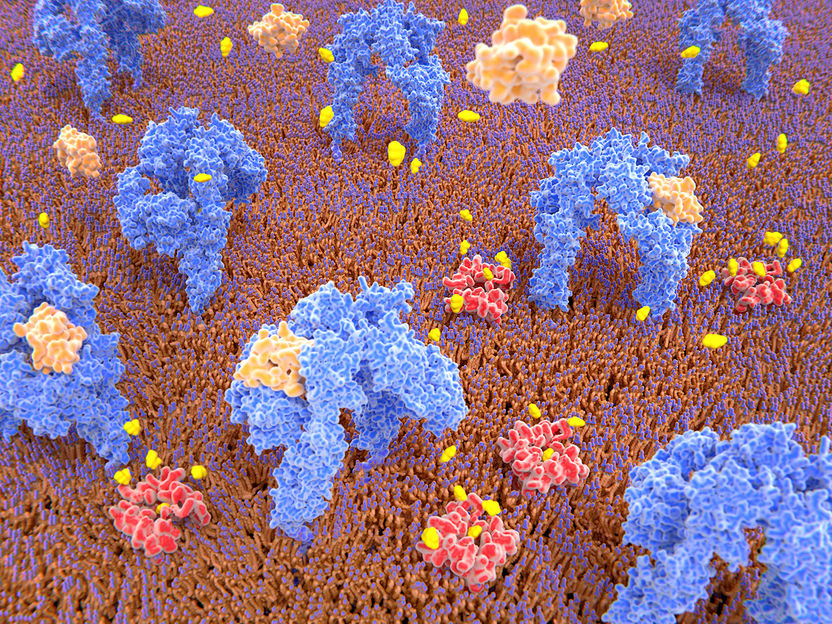
The new method Mass-Sensitive Particle Tracking (MSPT) allows the analysis of molecular mass, movement and interaction of proteins on biological membranes.
Juan Gärtner, AdobeStock
Cellular processes on membranes are often fast and short-lived. Molecules assemble briefly, separate again, interact with different partners and move along or through the membrane. It is therefore important to not only study static snapshots of these processes, but also to understand their dynamics. But how can this be achieved methodically? Petra Schwille from the MPIB and Nikolas Hundt from the LMU together with their team have developed the method Mass-Sensitive Particle Tracking - MSPT, which allows to analyze proteins during dynamic processes on membranes.
Analysis of dynamic processes on biological membranes
The starting point for the biophysicists were recent advancements in mass photometry, which could already be used to determine the molecular mass of unlabeled molecules in solution. What is new about MSPT is that the dynamics of membrane-associated proteins can now be tracked in their biologically plausible environment. In this process, individual proteins are identified by their molecular mass without the need for labeling. Frederik Steiert, one of the first authors of the publication, says: "We can now track directly on biological membranes what mass individual proteins have, how they move and how they interact. This allows us to study the dynamics of biological systems in greater detail." Analyzing dynamic processes is particularly important in biology as many processes at the membrane are transient.
Mass determination by light scattering
What principles is the new method based on? When light hits a particle, the light is scattered. The intensity of the scattered light depends on the mass of the particle. Videos in which individual proteins on membranes are made directly visible are recorded with a microscope. With the aid of analysis software, these proteins can be tracked and their scattering signal, and thus their mass, can be determined. This is currently possible for proteins with a molecular weight of at least 50 kDa, i.e. for a large part of all known proteins. Another advantage of the new MSPT method is that proteins do not have to be labeled. Labeling can be achieved, for example, by attaching fluorescent tags to molecules. However, labeling poses the risk that proteins could be impaired in their function or that the fluorescent labels could bleach during the experiment. By using MSPT, in contrast, methodological problems that can arise from labeling are prevented.
MinDE protein system
To demonstrate the potential of the method for biological questions, the biophysicists used an established system from the Schwille laboratory: the MinDE protein system from the bacterium Escherichia coli (E. coli). MinD and MinE proteins are involved in E. coli cell division. Tamara Heermann, another first author, says: "The method permits us to characterize properties of dynamical systems that were previously not measurable. This allowed us not only to verify established findings about the Min system, but also to gain new insights.” By using MSPT, the team was able to show that the complexes of MinD proteins are larger than initially thought. In addition, the experiments provide first insights that MinE can act as a connecting piece for MinD proteins and that it can thus initiate the membrane release of larger complexes.
As reported in the new Nature Methods paper, MSPT provides valuable insights for elucidating dynamic processes at biological membranes. However, the researchers are continuously working on improving the method even further. In the future, the method should also be applicable for integral membrane proteins and it should allow the detection of even smaller proteins.
Original publication
Other news from the department science
Most read news
More news from our other portals
See the theme worlds for related content
Topic world Protein analytics
Protein analytics provides a deep insight into these complex macromolecules, their structure, function and interactions. It is essential for discovering and developing biopharmaceuticals, understanding disease mechanisms, and identifying therapeutic targets. Techniques such as mass spectrometry, Western blot and immunoassays allow researchers to characterize proteins at the molecular level, determine their concentration and identify possible modifications.

Topic world Protein analytics
Protein analytics provides a deep insight into these complex macromolecules, their structure, function and interactions. It is essential for discovering and developing biopharmaceuticals, understanding disease mechanisms, and identifying therapeutic targets. Techniques such as mass spectrometry, Western blot and immunoassays allow researchers to characterize proteins at the molecular level, determine their concentration and identify possible modifications.