Scientists crack structure of a novel enzyme linked to cell growth and cancer
RNA, or ribonucleic acid, is present in the cells of all living beings and required to synthesize proteins. A research team at the University of California, Riverside, has discovered the structure of a novel RNA-modifying enzyme, ZCCHC4, and identified the mechanism that controls how this enzyme recognizes itssubstrate.
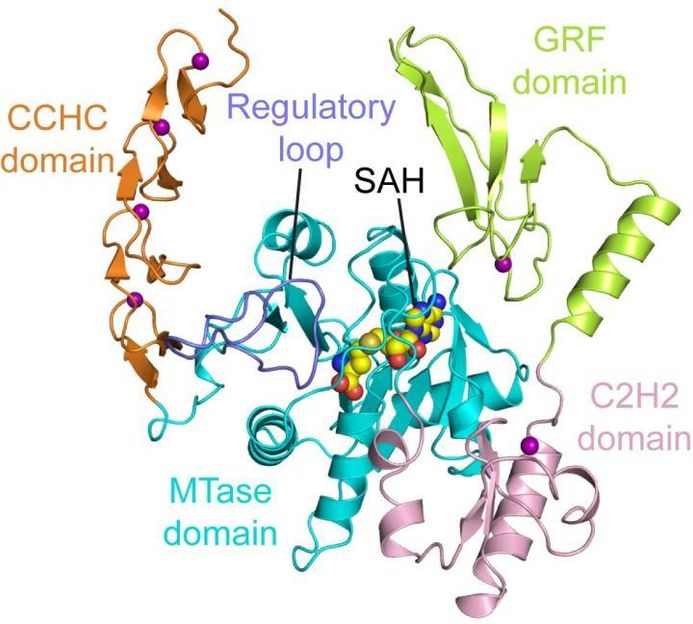
This image shows the structure of ZCCHC4.
Song lab, UC Riverside
ZCCHC4 influences cell proliferation and has been linked to cancers. It uniquely introduces one kind of RNA modification, N6-methyladenosine (m6A), into ribosomes, which are cell organelles made up of RNA molecules and protein.
The study, published in Nature Communications, explains how protein machineries in cells are regulated to target RNA molecules for m6A modification.
Jikui Song, an associate professor of biochemistry at UC Riverside who led the study, explained ZCCHC4 controls protein synthesis and cell proliferation by introducing an m6A modification into ribosomes. ZCCHC4, he added, is overexpressed in tumors associated with hepatocellular carcinoma -- the most common type of primary liver cancer.
"This is the first time anyone has determined the crystal structure of ZCCHC4," Song said. "Our discovery can be used for structure-based drug design against cancers and lead to a better understanding of how m6A, a modification associated with numerous biological processes, is installed on ribosomal RNA."
The m6A modification has received enormous attention in recent years due to the important role it plays in RNA metabolism and biology. How this modification is dynamically programmed and distributed in cells, however, remains poorly understood.
"The structure of ZCCHC4 provides an understanding of how this enzyme is wired to specifically act on '28S ribosomal RNA,'" Song said, noting a ribosome is assembled with differently sized subunits. 28S ribosomal RNA refers to the RNA component in the 28S ribosomal subunit. "We now understand that this enzyme is controlled by an 'autoinhibitory' mechanism that has been observed in many other cellular processes."
To crack the structure of ZCCHC4, Song's team first produced an enzymatically active and structurally rigid ZCCHC4 fragment. The researchers then coaxed this protein to crystallize. Finally, they diffracted the crystals using X-rays and analyzed the data, which led to the eventual discovery of ZCCHC4's structure.
Last year, Song's lab solved the crystal structure for an enzyme that plays a key role in DNA methylation, the process by which methyl groups are added to the DNA molecule.
Next, the research team will continue to explore how various DNA and RNA modifications in cells are created, which has strong implications in health and diseases.
Original publication
Most read news
Other news from the department science

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.