Getting to the heart of an enzyme
Decoding the heart of NatC: X-rays help shed light on structure
A team led by Oliver Daumke at the MDC has determined the three-dimensional structure of the NatC acetyltransferase in an article published in Nature Communications. This enzyme modifies cell proteins to enable them to perform their functions correctly. In cancer cells, however, NatC is often overactive.
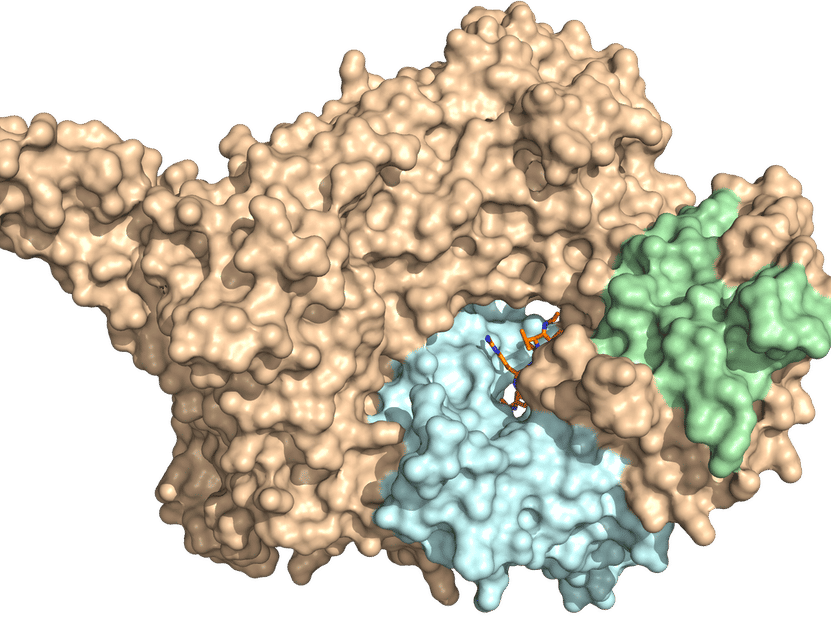
X-ray crystal structure of NatC. The three-part complex consists of a catalytic subunit (light blue) and a large and a small auxiliary subunit (brown and green, respectively). A prominent NatC tip is required for ribosome interaction.
© AG Daumke, MDC
Small changes can make a big difference. And this is also true of proteins: If just one hydrogen atom at the end of these huge molecules is swapped out for an acetyl group, this new chemical structure gives the protein completely different properties. It may now, for example, be able to interact with other proteins, its lifespan may change, or it may suddenly be able to seek out completely new targets in the cell.
An important cellular regulatory mechanism
Protein acetylation, which is the scientific term for the exchange of a hydrogen atom for an acetyl group, is therefore an important regulatory mechanism of cells. Acetylation can be set in motion by a group of enzymes called the N-terminal acetyltransferases, or NATs for short. These enzymes are acetylating the N-terminus of a protein, where an amino acid with a free amine group is located.
There are a large number of different NATs in human cells, but until recently, we only knew the detailed structure of a few of them. The precise structure of NatC acetyltransferase has now been “decoded” by a team led by Professor Oliver Daumke, head of the Structural Biology Lab at the Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC), together with researchers from Charité – Universitätsmedizin Berlin.
Most proteins in humans are acetylated
The study’s lead author is structural biologist Dr. Stephan Grunwald, a former PhD student and now a postdoc in Daumke’s team. “N-terminal acetylation is a very common protein modification – and thus extremely important for the many cellular functions,” says Grunwald. “In humans, some 80 to 90 percent of all proteins are modified this way.”
Grunwald started by taking bacteria of the species Escherichia coli (E. coli), and introduced into them the three NatC subunits. “These bacteria produced great quantities of NatC for us, which we then purified, crystallized, and examined more closely using X-ray crystallography,” reports Grunwald. It was already known that NatC consists of one catalytic and two auxiliary subunits. “But we first had to find out how these subunits are arranged in the NatC complex, and what functions each of them serves,” explains Grunwald.
X-rays help shed light on structure
For X-ray crystallography, it is necessary to first crystallize the enzyme by mixing it with particular chemical additives. “You end up with crystals about a tenth of a millimeter long, each containing many million NatC complexes that are arranged quite uniformly,” says Grunwald. The crystals are then flash-frozen at –196 degrees Celsius, after which they are bombarded with intense X-rays produced by a particle accelerator – such as the BESSY II facility in Berlin-Adlershof.
“Based on the diffraction pattern of the X-rays, we can draw conclusions about the three-dimensional structure of the protein,” explains Daumke. In doing so, he and his team discovered that the catalytic subunit of NatC, where the acetylation of proteins takes place, is surrounded by one small and one very large auxiliary subunit. “The latter serves as a kind of scaffold for the entire enzyme,” says Daumke.
The enzyme waits at the end of the production line
The researchers were also able to uncover other acetyltransferase secrets. “We already knew that NatC attaches itself to the ribosomes, which are the cell’s protein factories,” Grunwald reports. “With the help of binding assays, we could show that a positively charged NatC tip region is required for ribosome binding. It sits there, almost like it’s sitting at the end of a production line, and waits for a new, not-yet-folded protein to be rolled out.” NatC can tell from the first four amino acids of the newly produced protein whether it should acetylate it or not.
Decoding the heart of NatC
With the help of this new structural knowledge, the researchers were even able to gain detailed insight into the “beating heart” of NatC, the catalytic subunit. “We changed individual amino acids several times in the catalytic center of the enzyme,” Grunwald reports. He and his colleagues therefore now know exactly what the NatC heart looks like and the mechanism by which acetylation occurs there.
Blocking the catalytic center of NatC
This knowledge could one day be important for cancer therapy, for example. “NatC is often overactive in certain malignant cells,” says Grunwald. “So it is possible, at least hypothetically, to develop drugs that block the catalytic center of the enzyme – and thus also stop tumor growth.” First, however, the structural biologist wants to pass on his findings to cell biologists: “Now we have determined the exact structure of NatC, they can find out the precise functions of the enzyme in the human body.”