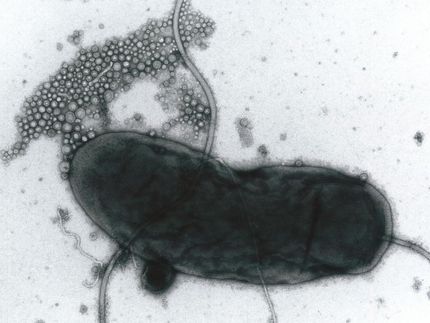
Detailed chemical structure of P22 virus resolved
Scientists at Baylor College of medicine, the Lawrence Berkeley National Laboratory, Massachusetts Institute of Technology and Purdue University have completed a model of unprecedented near-atomic resolution of the chemical structure of the P22 virus.
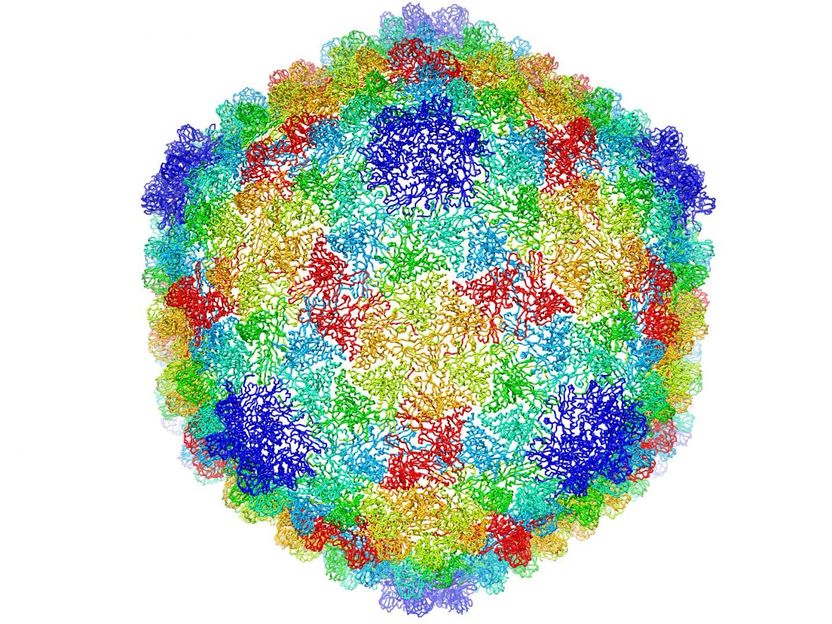
Complete capsid of bacteriophage P22 generated with validated atomic models that were derived from the high-resolution cryoEM density map.
C.Hryc and the Chiu Lab
For nearly 30 years the laboratory of Dr. Wah Chiu, Distinguished Service Professor and Alvin Romansky Professor of Biochemistry and Molecular Biology at Baylor and senior author of the paper, has been applying electron cryomicroscopy and computer reconstruction techniques to determine the 3-D structures of biological nanomachines, such as the P22 virus. This virus is a bacteriophage -- a type that infects bacteria, in this case Salmonella -and has been extensively studied through genetics, biochemistry and biophysics. Nevertheless, its precise chemical structure has remained unresolved.
"In 2011, we published a structure of the P22 virus that allowed us to trace out a majority of the protein backbone with certainty, but we could not visualize the fine details, such as individual, small side chains," said co-first author Corey Hryc, graduate student in the Chiu lab. "Since then, the technology in the microscopes has improved; we have new detectors that allow us to record better- and higher-contrast images to improve the resolution of our data. In addition, we have new processing algorithms that allow us to increase our ability to resolve the structure."
A new era of precision cryo-electron microscopy
"The novelty of this work is that we took more than 20,000 two-dimensional individual images of the P22 virus with the electron cryomicroscope and combined them using computational protocols to produce a 3-D map with unprecedented detail," Chiu said.
"By comparison, the two-dimensional images we take of the virus would be like taking thousands of photos of your head randomly, in different places, and then fit the photos where they would belong in a 3-D space using a computer," Hryc said. "In this way, we could probably create a 3-D model of your head, and, if the photos were X-ray images, we could see the insides, too."
The analysis of the high-resolution map of the P22 virus allowed the researchers to see in great detail all the building blocks of the proteins in the virus, the amino acids, including their side chains and how they interact with neighbor amino acids.
"The minute details we obtain now of biological structures with this approach give us more information about their biochemistry than before," Hryc said. "I think that's exciting."
"Thanks to this exquisite structural detail, we have determined the protein chemistry of the P22 virus," Chiu said. "I think it is important that we provide detailed annotations with the structure so other researchers can use it for their future experiments."
"Without the annotations you would think that everything in the map is essentially equivalent from a modeling standpoint, but this is not the case," Hryc said. "Some of these amino acids are much more clearly visible than others, particularly in the interactions between the different components of the virus."
"Hryc has set the standard for precision cryo-electron microscopy," Chiu said.
Original publication
Original publication
Corey F. Hryc, Dong-Hua Chen, Pavel V. Afonine, Joanita Jakana, Zhao Wang, Cameron Haase-Pettingell, Wen Jiang, Paul D. Adams, Jonathan A. King, Michael F. Schmid, and Wah Chiu; "Accurate model annotation of a near-atomic resolution cryo-EM map"; PNAS; 2017
Topics
Organizations
Other news from the department science

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.