Breakthrough in protein research: Toolkit makes protein design faster and more accessible
What used to take weeks can now be completed in a day or less
Researchers at the Max Planck Institute for Biology Tübingen, the University of Tübingen, and the University Hospital Tübingen have developed a web-based toolkit to accelerate and simplify protein design without needing powerful computers or extensive protein design expertise on the user’s end. The toolkit benefits its users with multiple design tools, fast analyses, easy interpretation, and downloadable results. Their framework, published in Nucleic Acids Research, offers a comprehensive resource for biological research community.

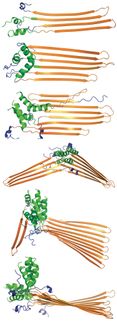
The Damietta Server seamlessly integrates different protein design tools and provides their interoperability. Image adapted from Nucleic Acids Res, gkae297, https://doi.org/10.1093/nar/gkae297
© Dr. Kateryna Maksymenko / Max-Planck-Institut für Biologie Tübingen
Designing proteins with novel functions is a crucial step in many areas of biomedical research. Protein design process has often been complex, requiring significant computing power and expertise, thus limiting its applicability and accessibility to a broader range of researchers.
The Damietta protein design toolkit addresses these challenges by offering a web-based platform that integrates several protein design tools. Such a platform was inspired by the MPI bioinformatics toolkit, which greatly simplifies bioinformatics workflows, and is hosted by the Department of Protein Evolution at the Max Planck Institute for Biology Tübingen, headed by Prof. Andrei Lupas, who supported the development of the new protein design toolkit.
“Damietta’s user-friendly interface makes it a powerful tool for researchers of all experience levels," says Dr. Kateryna Maksymenko, a researcher in the Department of Protein Evolution at the Max Planck Institute for Biology Tübingen. "This toolkit significantly accelerates protein design, boosting discoveries and direct clinical applications in biomedicine."
Unlike machine learning-based methods, Damietta relies on well-established physics principles, ensuring generalisable, accurate, and interpretable results.
The Damietta Server simplifies the process of protein design for users by offering:
- Faster and more accurate calculations. What used to take weeks can now be completed in a day or less.
- Intuitive graphical interface, allowing researchers with limited protein design experience to use the toolkit effectively.
- Improved transparency by providing clear and interpretable results, enabling researchers to understand the underlying physical interactions influencing the designed proteins.
Wider potential for new biomedical and therapeutic applications
Previously, Damietta software demonstrated its effectiveness by designing two classes of proteins for cancer treatment. The team also uses the software to develop proteins that are helpful for various disorders, including bone marrow failure and leukaemia.
Current users can look forward to future updates, as the researchers are actively developing Damietta to enable setting up membrane environment and to incorporate non-standard amino acids and small molecules.
Original publication
Other news from the department science

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents

DNA analysis reveals risk of post-meal insulin resistance - Targeted intervention may be possible
OGT Grants DNA Sequence Variation Licence to Biotype Diagnostics GmbH

New Method for Analysing Nanoporous Materials - Innovative method opens up new possibilities for materials science

A milestone on the pathway to Lab 4.0: A new standard for the smart lab - SPECTARIS presents the first industrial communication standard for laboratory and analytical devices

Fish and seafood - improved trace detection of life-threatening allergy sources - "AQUALLERG-ID": Researchers develop methods for detecting potential food allergens
PerkinElmer appoints President & Chief Operating Officer
ECHA calls for information to avoid unneccessary animal testing
Newly improved NIST reference material targets infant formula analysis

LAUDA appoints Dr. Ralf Hermann as the new General Manager Constant temperature equipment

A relative from the Tianyuan Cave - Ancient DNA has revealed that humans living some 40,000 years ago in the area near Beijing were likely related to many present-day Asians and Native Americans
Measurable for the first time: How bio molecules react to lack of space - Sensor demonstrates lack of space in living cells