Human environmental genome recovered in the absence of skeletal remains
Innovative approach permits the identification of DNA in samples of environmental material
Ancient sediments from caves have already proven to preserve DNA for thousands of years. The amount of recovered sequences from environmental sediments, however, is generally low, which difficults the analyses to be performed with these sequences. A study led by Ron Pinhasi and Pere Gelabert of the University of Vienna and published in Current Biology successfully retrieved three mammalian environmental genomes from a single soil sample of 25,000 years bp obtained from the cave of Satsurblia in the Caucasus (Georgia).

Overview of the excavation works of Satsurblia cave in 2017
© Anna Belfer-Cohen
The cave of Satsurblia was inhabited by humans in different periods of the Paleolithic: Up to date a single human individual dated from 15,000 years ago has been sequenced from that site. No other human remains have been discovered in the older layers of the cave.
The innovative approach used by the international team led by Prof. Ron Pinhasi and Pere Gelabert with Susanna Sawyer of the University of Vienna in collaboration with Pontus Skoglund and Anders Bergström of the Francis Crick Institute in London permits the identification of DNA in samples of environmental material, by applying extensive sequencing and huge data analysis resources. This technique has allowed the recovery of an environmental human genome from the BIII layer of the cave, which is dated before the Ice Age, about 25,000 years ago.
This new approach has evidenced the feasibility of recovering human environmental genomes in the absence of skeletal remains. The analysis of the genetic material has revealed that the SAT29 human environmental genome represents a human extinct lineage that contributed to the present day West-Eurasian populations. To validate the results, the researchers compared the recovered genome with the genetic sequences obtained from bone remains of the nearby cave of Dzudzuana, obtaining definitive evidence of genetic similarities. This fact validate the results and excludes the possibility of modern contamination of the samples.
Along with the identified human genome, other genomes such as wolf and bison have also been recovered from the environmental samples. The sequences have been used to reconstruct the wolf and bison Caucasian population history and will help better understand the population dynamics of these species.
The team now plans to perform further analyses of soil samples from the cave of Satsurbia with the objective of revealing interactions between extinct fauna and humans and the effect of climatic changes on mammalian populations. The ability to recover DNA from soil samples allows us the reconstruction of the evolution of whole past ecosystems .
Original publication
Other news from the department science

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents

Yumda now part of LUMITOS - Europe’s leading provider of B2B portals has expanded its portfolio to media outlets for the food and beverage industries

Analytica conference 2020 for the first time virtual - Topics in focus: Antibiotic resistance and the “laboratory of the future”

Schizophrenia manifests itself in the brain structure - The symptoms of schizophrenia vary greatly from person to person
The race is on for new lithium ion batteries - Battery consortium headed by BASF receives € 21 million sponsorship from German Ministry of Education and Research
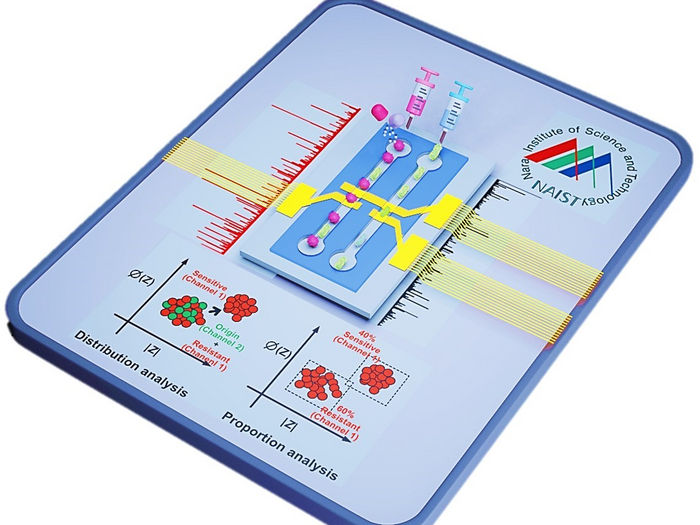
Antibiotic resistance testing no longer impeded by time - Researchers have developed a novel method for antibiotic resistance testing that might transform microbial screening in clinical and research labs
A new way to stimulate cellular recycling process

Thermo Fisher Scientific and QIAGEN N.V. Agree on Amended Terms to Acquisition Agreement - Offer price increased

New insights into the function of a risk factor for Alzheimer’s disease - Researchers decipher the structure-function relationship of the multifunctional protein clusterin for the first time

Rigaku opens Rigaku Technology Center Taiwan - Advancing regional growth through engineering and collaboration in Taiwan and beyond