Non-coding RNA reveals its secrets
Scientists from the Schuman Lab at the Max Planck Institute for brain Research have investigated the variety of structures and functions in non-coding parts of messenger RNA (mRNA) from brain cells. These untranslated regions (UTRs) are essential for regulating the stability and localization of mRNAs, as well as its translation into proteins.
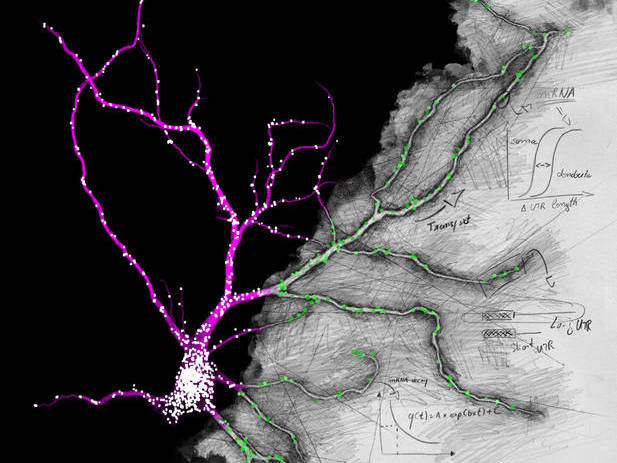
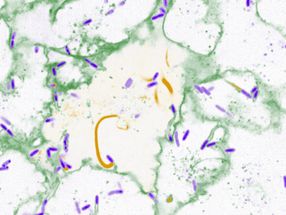
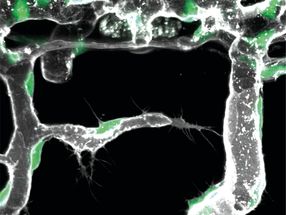
Fluorescence in situ hybridization against the unique sequence of the long 3’ untranslated region of Camk2a in a hippocampal neuron. Neurons localize mRNAs near synapses where their translation can be regulated by synaptic demand and activity. Tushev, Glock et al. discover a huge diversity in neuronal mRNA 3’UTRs, which give rise to differences in localization, stability, translation, and plasticity.
Artwork by Maximilian Heumüller.
Our brain’s capacity to learn and to form memories relies on the ability to respond adaptively to environmental inputs. The integration of these inputs occurs at synapses, the connections between brain cells. Unlike other cell types in the body, neurons have an elaborate architecture, consisting of axons to transmit and dendrites to receive information. Like all cells, neurons use proteins to carry out their important cellular functions. Neurons supply their processes with proteins by moving mRNA molecules to the dendrites and axons where they can be locally translated.
A mRNA molecule consists of a coding sequence in the middle, which serves as a template for new proteins, and two non-coding regions located on both ends: the 3’ region on the right and 5’ on the left hand side. These so-called untranslated regions (UTRs) control gene expression at the RNA level and are variable in length and composition. By acting as binding platforms, they can regulate stability, trafficking and translation, thus dictating the fate of an mRNA molecule within a cell.
Researchers from the Schuman Lab now sequenced the mRNA present in different regions of rat hippocampal slices and took a closer look at the 3’ UTR region. Schuman: “When we examined mRNA in both neuronal cell bodies and in the neuropil, primarily consisting of axons and dendrites, we noticed a huge diversity in the 3’UTRs of neuronal mRNAs with many transcripts showing enrichment for a particular 3’UTR isoform in a cellular compartment. In addition, the isoforms enriched in the neuropil proved to be more stable than those located in the cell body.” The scientists were even able to follow the dynamics of these molecules during a period of enhanced neural activity and observed an alteration of 3’UTR isoforms present in each compartment. The Schuman team speculates that some of the changes in the dendritic compartment might be due to local remodeling of 3’UTRs.
Original publication
Original publication
Georgi Tushev, Caspar Glock, Maximilian Heumüller, Anne Biever, Marko Jovanovic, Erin M. Schuman; "Alternative 3′ UTRs Modify the Localization, Regulatory Potential, Stability, and Plasticity of mRNAs in Neuronal Compartments"; Neuron; 2018
Topics
Organizations
Other news from the department science

Get the analytics and lab tech industry in your inbox
From now on, don't miss a thing: Our newsletter for analytics and lab technology brings you up to date every Tuesday. The latest industry news, product highlights and innovations - compact and easy to understand in your inbox. Researched by us so you don't have to.